


3.2. The atomic data input file
The user must supply all necessary atomic data used by PopRatio in a separate input file, that will be read by INITPOPRATIO. We provide input files containing atomic data pertaining five atoms/ions of interest in fine structure absorption line diagnosis of QSO absorbers: C0, C+, Si+, O0 and Fe+. Although in these files we list all the references in the literature where the atomic data were taken, we do not discuss the particular models adopted here, since this will be the subject of a forthcoming paper (Silva and Viegas, in preparation).
Next we describe the general syntax of the input file. As an example we take the file OI.dat, that contains the atomic data of neutral oxygen.
The atomic data information is distributed along several blocks, each one starting with the "#" character. The user is free to insert any comments before this symbol. The first block:
#
OI
is the name of the atom/ion being considered. This string is stored in the global variable SPECIES_NAME. It may be up to 10 characters long. The next blocks of data are:
#
3
this is the number of levels that belong to the ground term. In atomic oxygen these levels are 3P2, 1, 0. PopRatio will take into account fluorescence for transitions among these levels. This value is stored in the global variable NGROUND.
#
5
this is the maximum number of levels that may be taken into account. The user must make sure to enter enough data to allow PopRatio to consider this number of levels. If INITPOPRATIO is called with a value for NBLEVELS higher than this, a warning message will be issued.
| # | |||
| 0.0 | 5 | 1 | 2s2 2p4 3P2 |
| 158.265 | 3 | 2 | 2s2 2p4 3P1 |
| 226.977 | 1 | 3 | 2s2 2p4 3P0 |
| 15867.862 | 5 | 4 | 2s2 2p4 1D2 |
| 33792.583 | 1 | 5 | 2s2 2p4 1S0 |
these are the values for the energies and statistical weights for the levels. The first column is the energy relative to the ground level (in cm-1), and the second is the statistical weight for the level. The last two columns numbering and naming the levels are not read by INITPOPRATIO, they are just entered for user reference. The energies must be sorted in ascending order. The energies and statistical weights are stored in global variables E(i) and G(i), respectively.
| # | ||
| 9 | ||
| 1 | 2 | 8.865E-5 |
| 1 | 3 | 1.275E-10 |
| 2 | 3 | 1.772E-5 |
| 1 | 4 | 6.535E-3 |
| 2 | 4 | 2.111E-3 |
| 3 | 4 | 6.388E-7 |
| 1 | 5 | 2.945E-4 |
| 2 | 5 | 7.909E-2 |
| 4 | 5 | 1.124E0 |
This block contains the transition probabilities. The number at the beginning is the number of transitions listed. The first column is the lower level index j for the transition, next are the higher level index i and the transition probability Aij (in s-1). The transition probabilities are stored in the global variable A(i,j). As the transition probabilities are read, INITPOPRATIO automatically sets the Einstein coefficients by eq. (3) and stores them in the global variable B(i,j). There is no need to enter null transition probabilites, since transition probabilities not entered are assumed to be zero.
| # | |||
| 135 | |||
| 76794.798 | 3 | 1 | 3.30E+08 |
| 76794.798 | 3 | 2 | 1.97E+08 |
| 76794.798 | 3 | 3 | 6.54E+07 |
| 96225.049 | 3 | 1 | 9.31E+07 |
| 96225.049 | 3 | 2 | 5.56E+07 |
| 96225.049 | 3 | 3 | 1.85E+07 |
| 97488.378 | 3 | 1 | 1.99E+06 |
| 97488.378 | 3 | 2 | 2.97E+07 |
| 97488.378 | 3 | 3 | 3.95E+07 |
| 97488.448 | 5 | 1 | 1.79E+07 |
| 97488.448 | 5 | 2 | 5.35E+07 |
| ............................................................. | |||
These are the transitions involving the µ higher-lying
levels of section 2.2 and the NGROUND ground
term levels. The number
at the beginning is
the number of transitions listed. The first column is the energy
relative to the ground level
(in cm-1) of the higher-lying level, the second is its
statistical weight, the third is
the lower-lying level index (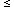 NGROUND) for the transition and the fourth is the transition
probability (in s-1). The energy levels must be entered in
ascending order.
If some higher-lying level was already loaded in by INITPOPRATIO in the
fourth block (and
therefore will be explicitly included in the system of statistical
equilibrium equations),
then it will not be taken into account when POPULATIONRATIO calculates
the indirect excitation
rates, GAM(i,j). However its contribution will be added in TOTGAM(i,j),
in case the user wishes to assess it.
NGROUND) for the transition and the fourth is the transition
probability (in s-1). The energy levels must be entered in
ascending order.
If some higher-lying level was already loaded in by INITPOPRATIO in the
fourth block (and
therefore will be explicitly included in the system of statistical
equilibrium equations),
then it will not be taken into account when POPULATIONRATIO calculates
the indirect excitation
rates, GAM(i,j). However its contribution will be added in TOTGAM(i,j),
in case the user wishes to assess it.
#
4
This is the number of collision partners. This value is stored in the global variable NPARTNERS and is the dimension of the collisional data type vector CD(i).
Next we have one block for each one of the collisional partners:
| # | ||||||||
| electron | ||||||||
| 5 | ||||||||
| 10 | ||||||||
| 3 | 7 | |||||||
| 50. | 100. | 200. | 500. | 1000. | 2000. | 3000. | ||
| 2 | 1 | 8.34E-04 | 1.26E-03 | 1.79E-03 | 2.58E-03 | 3.35E-03 | 4.61E-03 | 5.92E-03 |
| 3 | 1 | 3.23E-04 | 5.00E-04 | 7.34E-04 | 1.12E-03 | 1.49E-03 | 2.08E-03 | 2.64E-03 |
| 3 | 2 | 2.74E-07 | 8.92E-07 | 2.78E-06 | 1.05E-05 | 2.38E-05 | 4.62E-05 | 6.98E-05 |
| ............................................................................................................................................... | ||||||||
The first line is the collisional partner name, with a maximum of 10 characters long. This string is stored in the global variable CD(i)%PARTNER_NAME. The second line is the index of the highest-lying level that may be populated by collisions with this partner; it is stored in CD(i)%MAXLEVEL. The third line is the total number of transitions listed below; it is stored in CD(i)%NINTERPOL. For each transition we have a table of points containing the collision rates (or Maxwellian-averaged collision stregths, if the partner name is 'ELECTRON') as a function of the kinetic temperature. PopRatio interpolates this table of points by a cubic-spline in log-log space (linear-log, in case the partner name is 'ELECTRON'). PopRatio also uses eq. (6) to calculate the inverse process rate.
In the next line we have the partial number of transitions that follows and the number of data points entered in the tables. The next line lists the temperature values (in K) for the transitions that follows. Then we have the collision rates (in cm3s-1, or Maxwellian-averaged collision strengths), the first two values entered giving the i and j indexes for the transitions ( i -> j). If the partner name is electron then the order of these indexes are irrelevant, otherwise i must come first. This is repeated until the sum of the partial number of transitions equals the total number of transitions listed. This is to allow greater flexibility, since rates for the various transitions might be taken from different sources in the literature and therefore have been calculated in different temperature values.
| # | |||
| proton | |||
| 3 | |||
| 3 | |||
| 3 | 2 | ||
| 1.E30 | 1.E30 | ||
| 2 | 1 | 1.E-30 | 1.E-30 |
| 3 | 1 | 1.E-30 | 1.E-30 |
| 3 | 2 | 1.E-30 | 1.E-30 |
This is an example of how the user can implement an analytical fitting. The transitions induced by collisions with protons are entered, but with an unphysical temperature range. Since the temperature passed to POPULATIONRATIO will always lie outside this range, it will call RATE_EXTRAPOL that is then used to call OI_PROTON, containing the analytical fittings.
The last two blocks in OI.dat give the collision rates for transitions induced by collisions with neutral hydrogen and neutral helium.